 |
Predicting
gene sequences with AI to study codon
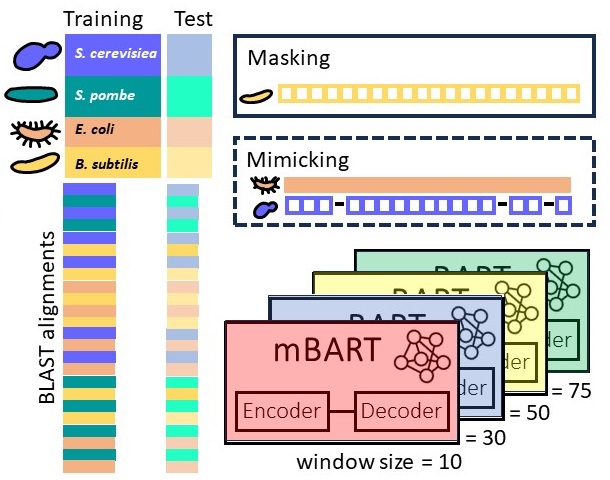
usage patterns We developed and trained an mBART model to predict codon sequences used by an organism to encode a given amino acid sequence. Our models improve upon frequency-based predictions using contemporary AI tools that learn complex patterns and capture interactions between codons. with Tomer Sidi, Shiri
Bahiri-Elitzur, and Tamir Tuller |
|
 |
CyToStruct - External Application Hooking for Cytoscape with Sergey Nepomnyachiy and Nir
Ben-Tal install,
Bitbucket
(including source code and demos) and slides
from a workshop Sergey did |
|
|
VISTAL - A two-dimensional visualization tool for structural alignments with Barry Honig download linux
executable, example
|
||